| DPOGS214587 | ||
|---|---|---|
| Transcript | DPOGS214587-TA | 348 bp |
| Protein | DPOGS214587-PA | 115 aa |
| Genomic position | DPSCF300050 - 418791-422534 | |
| RNAseq coverage | 526x (Rank: top 24%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | % | |||
| Bombyx | BGIBMGA005153-TA | 1e-27 | 83.33% | |
| Drosophila | CG7593-PA | 5e-10 | 37.33% | |
| EBI UniRef50 | UniRef50_E2C819 | 6e-13 | 48.65% | N-acetyltransferase 11 n=8 Tax=Formicidae RepID=E2C819_HARSA |
| NCBI RefSeq | XP_001601002.1 | 7e-13 | 47.30% | PREDICTED: hypothetical protein [Nasonia vitripennis] |
| NCBI nr blastp | gi|328788451 | 2e-14 | 49.35% | PREDICTED: n-alpha-acetyltransferase 40, NatD catalytic subunit-like [Apis mellifera] |
| NCBI nr blastx | gi|328788451 | 1e-11 | 49.35% | PREDICTED: n-alpha-acetyltransferase 40, NatD catalytic subunit-like [Apis mellifera] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0008152 | 1.8e-05 | metabolic process | |
| GO:0008080 | 1.8e-05 | N-acetyltransferase activity | ||
| KEGG pathway | dre:550325 | 5e-10 | ||
| K00680 (E2.3.1.-) | maps-> | Benzoate degradation via CoA ligation | ||
| Limonene and pinene degradation | ||||
| Ethylbenzene degradation | ||||
| Tyrosine metabolism | ||||
| 1- and 2-Methylnaphthalene degradation | ||||
| InterPro domain | [27-91] IPR016181 | 4.6e-09 | Acyl-CoA N-acyltransferase | |
| Orthology group | MCL30309 | Lepidoptera specific | ||
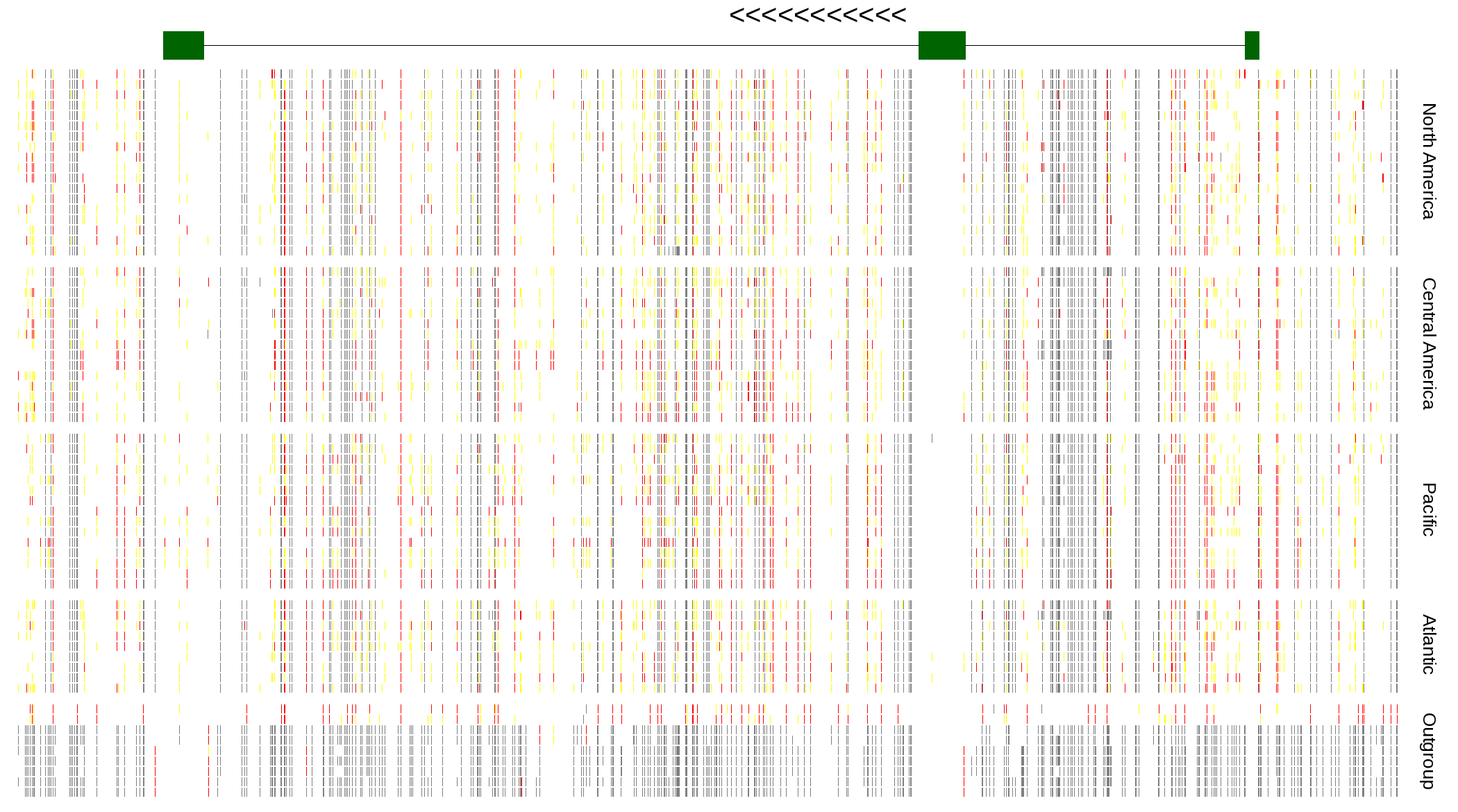
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS214587-TA
ATGTATCAATCCGACCTTATTCAGGTCAACGCATCTGCATTCCGTGTAAGCTATGAGGTTCAGATCGAGGAGGGCGGCCGTCGTCGCGGGCTTGGTCAGAGAGTGCTCTGTGTTCTGGAGCGACTGGCTCAGGCCACACGGATGAGGTGTGTCCGGCTCACAGCCCTCACGCACAACCCCTCGGCAGCTGCATTTTTCAGAGCGTGCGGGTACAGTATAGACGACACAAGCCCGCCCAAAGAGGAGACGAACCATTACGAGATACTCAGCAAGGCTACTGAACACGTGGAGGGTGAAGACCCCGAGGAACAGTCAAGAGATGGCGCTATACAAACAGACATACACTGA
>DPOGS214587-PA
MYQSDLIQVNASAFRVSYEVQIEEGGRRRGLGQRVLCVLERLAQATRMRCVRLTALTHNPSAAAFFRACGYSIDDTSPPKEETNHYEILSKATEHVEGEDPEEQSRDGAIQTDIH-