| DPOGS214899 | ||
|---|---|---|
| Transcript | DPOGS214899-TA | 567 bp |
| Protein | DPOGS214899-PA | 188 aa |
| Genomic position | DPSCF300135 - 364752-367691 | |
| RNAseq coverage | 16x (Rank: top 81%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL017186 | 1e-30 | 46.83% | |
| Bombyx | BGIBMGA012478-TA | 1e-08 | 26.67% | |
| Drosophila | epsilonTry-PA | 5e-11 | 25.30% | |
| EBI UniRef50 | UniRef50_G9F9I9 | 1e-09 | 27.22% | Seminal fluid protein CSSFP037 n=1 Tax=Chilo suppressalis RepID=G9F9I9_9NEOP |
| NCBI RefSeq | XP_002006761.1 | 3e-10 | 25.77% | GI21243 [Drosophila mojavensis] |
| NCBI nr blastp | gi|364023625 | 5e-09 | 27.22% | seminal fluid protein CSSFP037 [Chilo suppressalis] |
| NCBI nr blastx | gi|364023625 | 1e-08 | 26.16% | seminal fluid protein CSSFP037 [Chilo suppressalis] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0003824 | 4.3e-28 | catalytic activity | |
| GO:0004252 | 1.2e-15 | serine-type endopeptidase activity | ||
| GO:0006508 | 1.2e-15 | proteolysis | ||
| KEGG pathway | dpo:Dpse_GA15051 | 1e-08 | ||
| K01312 (E3.4.21.4, PRSS1, PRSS2, PRSS3) | maps-> | Neuroactive ligand-receptor interaction | ||
| InterPro domain | [1-170] IPR009003 | 4.3e-28 | Peptidase cysteine/serine, trypsin-like | |
| [11-165] IPR001254 | 1.2e-15 | Peptidase S1/S6, chymotrypsin/Hap | ||
| Orthology group | MCL35038 | Lepidoptera specific | ||
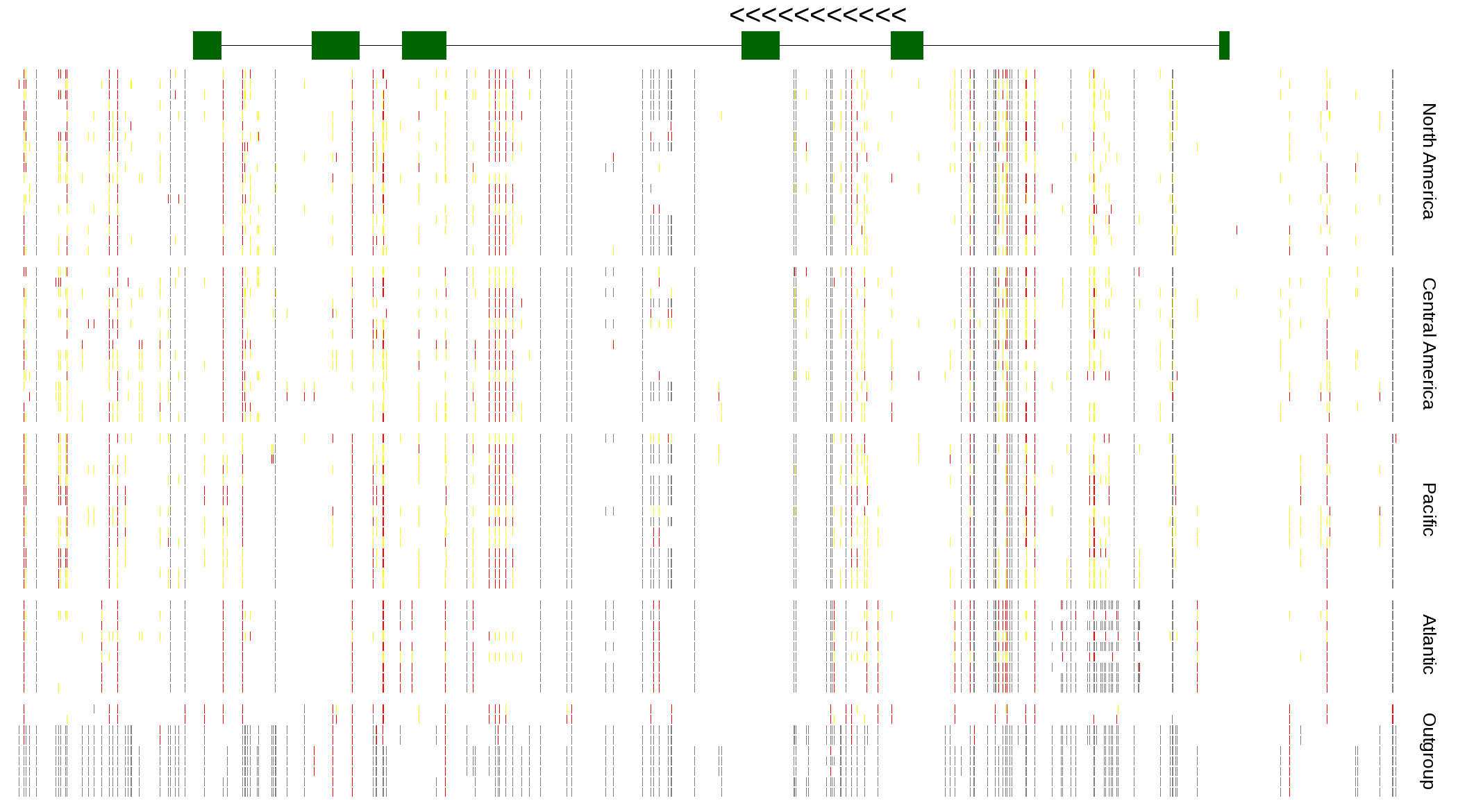
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS214899-TA
ATGGGCGCCAATACTATCACAGGGATTGGACAAGTAATACCCGTAGCAGAATATAAAATTCATGAGTACTTCAAACAGCTGAGCGCTCTCGACAATGATATTGCTTTGTTGGTGCTTCAAAAAAATGTAAAATATGAAACGACAGTTAAAAAAACACTCCTCGTTGAACCAGAGATTGCAATCCGAGAAAACACTAATCTCTTGGTCGCGGGATGGGGCGGCGGCACGCAATTGCCGATGAATTATATCAACTTACTCCTCCAATTAGGAGCTGTAGTTCTCAAGAGATCTGAATGCGTACAATATTATGGTCAATTGATGACGCCATCAAACTTCTGCGTTCGATACACAACGGATCACTTTTTAACTGACAATGGTGGCGGCGCCATGTTTGACGATCTACTAGTCGGTATATTATCTGTAGGAGAAATCAAAACCGATATTAGATTCAACACAGCAATAGTTACCAACGTGTCTTATTTCCACAGGTGGATTACACTCAACTCGAAAAGGTTCTTGAAAAAGTACTGTATAGTTCAGGATAAACCGAACATATCTCTCAAATGA
>DPOGS214899-PA
MGANTITGIGQVIPVAEYKIHEYFKQLSALDNDIALLVLQKNVKYETTVKKTLLVEPEIAIRENTNLLVAGWGGGTQLPMNYINLLLQLGAVVLKRSECVQYYGQLMTPSNFCVRYTTDHFLTDNGGGAMFDDLLVGILSVGEIKTDIRFNTAIVTNVSYFHRWITLNSKRFLKKYCIVQDKPNISLK-