| DPOGS215081 | ||
|---|---|---|
| Transcript | DPOGS215081-TA | 645 bp |
| Protein | DPOGS215081-PA | 214 aa |
| Genomic position | DPSCF300187 - 165819-171970 | |
| RNAseq coverage | 312x (Rank: top 36%) | |
| Annotation | ||||
|---|---|---|---|---|
| Heliconius | HMEL006736 | 6e-18 | 31.40% | |
| Bombyx | BGIBMGA007183-TA | 6e-36 | 42.71% | |
| Drosophila | Jon99Ci-PA | 6e-08 | 40.79% | |
| EBI UniRef50 | UniRef50_B6D1R2 | 3e-44 | 43.67% | Serine proteinase diverged n=1 Tax=Ostrinia nubilalis RepID=B6D1R2_OSTNU |
| NCBI RefSeq | XP_001653094.1 | 6e-08 | 27.85% | trypsin-alpha, putative [Aedes aegypti] |
| NCBI nr blastp | gi|304443627 | 4e-44 | 45.41% | serine protease 38 [Mamestra configurata] |
| NCBI nr blastx | gi|304443627 | 1e-43 | 45.41% | serine protease 38 [Mamestra configurata] |
| Group | ||||
|---|---|---|---|---|
| Gene Ontology | GO:0003824 | 3.3e-23 | catalytic activity | |
| GO:0004252 | 7.1e-13 | serine-type endopeptidase activity | ||
| GO:0006508 | 7.1e-13 | proteolysis | ||
| KEGG pathway | ||||
| InterPro domain | [10-213] IPR009003 | 3.3e-23 | Peptidase cysteine/serine, trypsin-like | |
| [25-208] IPR001254 | 7.1e-13 | Peptidase S1/S6, chymotrypsin/Hap | ||
| Orthology group | MCL25320 | Lepidoptera specific | ||
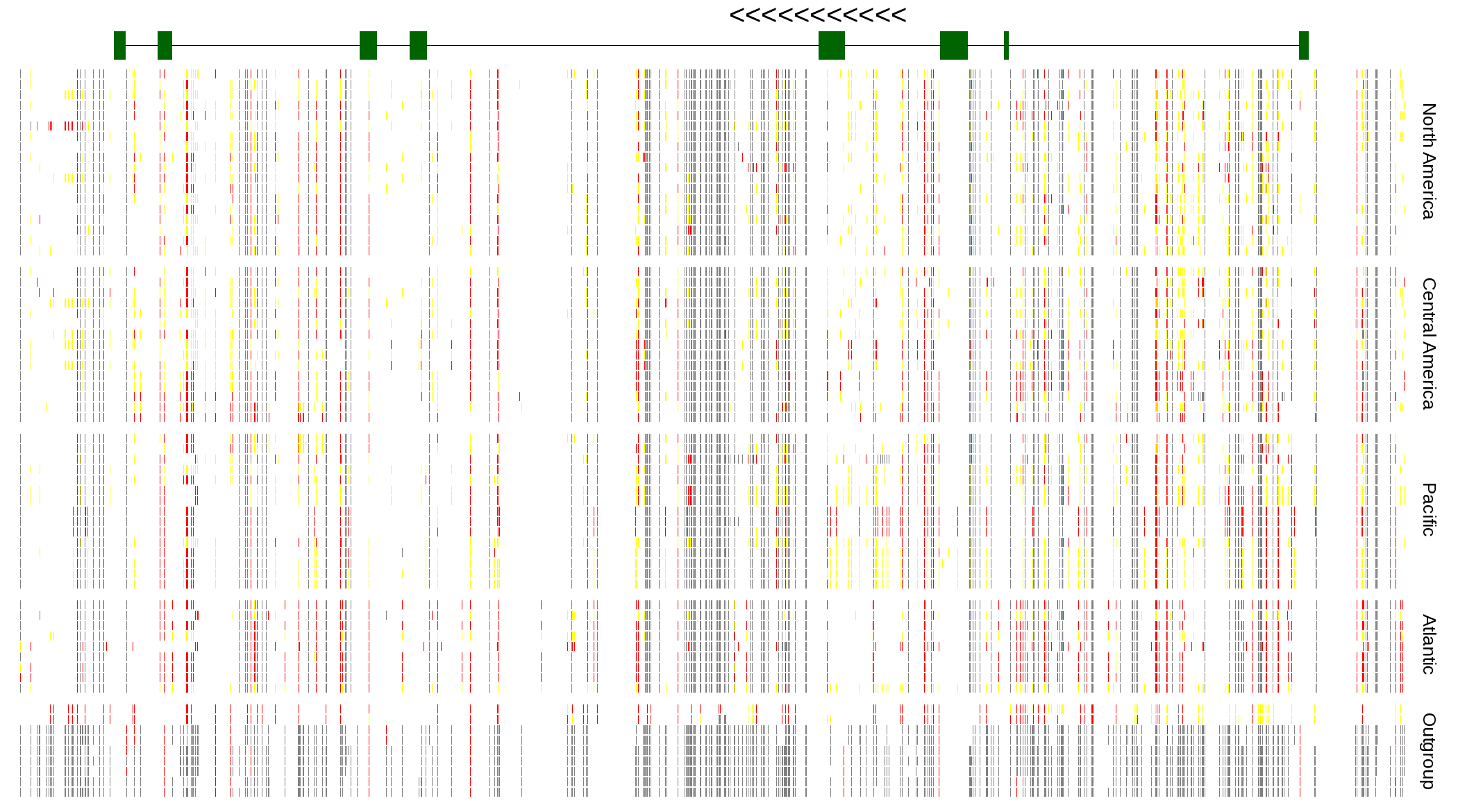
| Genotypes for resequenced monarchs and outgroup Danaus species |
|---|

>DPOGS215081-TA
ATGAAGGTTCTCGTTGTTGCTTTCCTGTGCACCTTCGCCGCCGTCCAGGGGCGTAGCGCATTGAGTGAATTGTCAGCGTCTGCTGGCGAGAATCCTTACGTGGTTCACATCCGCCTGGCGACTTCTGATAACAATGGACTTCTCAGCACATGCGCTGGCTCCCTGATCGGCTCCACATGGGTTCTTACAAGCGCTGACTGCGTTAAAAATTCTCGTTTCATGTGGCTTCGTTTCGGCGCTGTAGACGTGTTCCGCCCCGCGTTTTTGGCTGAGGGGAACCTCAACGTGCGCGTCCAAGATGGACTCGCTCTTATCAACGTGAACAGAGCCATCGAAGAGAACGACGTAATTCGTCGCGTGTCTTTAGCTGAGGCTGAATCCGAAGTTCCTGAAAGCGGCAAATTGTGCGTCTTCGGAGCTGAAAACGATGCACCCGGAAGTCAATTGAGTTGTTCAAACGTCGAGGTTGCTGCCAATGAAGCCGGCGAACTCGTAGCTGACAGTGGAGCTTCCAAATTCGACCTCGGAGCGGCTCTGGTGAGTGACGGCGTACAAGTTGGAGTACTTTCAAGCGTTGGCGAATCATCTGTGTTCGCATCAGTTGCCAAAAATAGAGATTGGATCAAACAAGTCACCGGTTTGTAA
>DPOGS215081-PA
MKVLVVAFLCTFAAVQGRSALSELSASAGENPYVVHIRLATSDNNGLLSTCAGSLIGSTWVLTSADCVKNSRFMWLRFGAVDVFRPAFLAEGNLNVRVQDGLALINVNRAIEENDVIRRVSLAEAESEVPESGKLCVFGAENDAPGSQLSCSNVEVAANEAGELVADSGASKFDLGAALVSDGVQVGVLSSVGESSVFASVAKNRDWIKQVTGL-